Computational multifocal microscopy
Published in Biomedical Optics Express, 2018
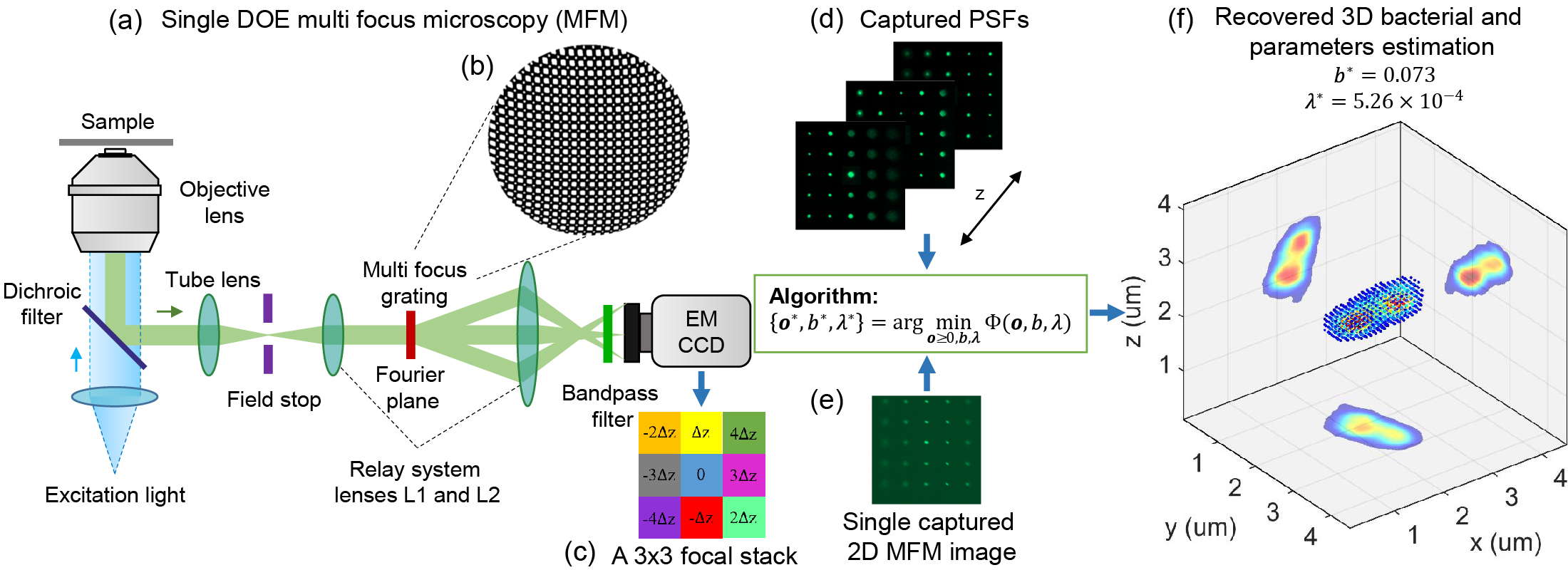
Abstract: Despite recent advances, high performance single-shot 3D microscopy remains an elusive task. By introducing designed diffractive optical elements (DOEs), one is capable of converting a microscope into a 3D “kaleidoscope,” in which case the snapshot image consists of an array of tiles and each tile focuses on different depths. However, the acquired multifocal microscopic (MFM) image suffers from multiple sources of degradation, which prevents MFM from further applications. We propose a unifying computational framework which simplifies the imaging system and achieves 3D reconstruction via computation. Our optical configuration omits optical elements for correcting chromatic aberrations and redesigns the multifocal grating to enlarge the tracking area. Our proposed setup features only one single grating in addition to a regular microscope. The aberration correction, along with Poisson and background denoising, are incorporated in our deconvolution-based fully-automated algorithm, which requires no empirical parameter-tuning. In experiments, we achieve spatial resolutions of 0.35um (lateral) and 0.5um (axial), which are comparable to the resolution that can be achieved with confocal deconvolution microscopy. We demonstrate a 3D video of moving bacteria recorded at 25 frames per second using our proposed computational multifocal microscopy technique.
Citation: Kuan He, Zihao Wang, Xiang Huang, Xiaolei Wang, Seunghwan Yoo, Pablo Ruiz, Itay Gdor, Alan Selewa, Nicola J. Ferrier, Norbert Scherer, Mark Hereld, Aggelos K. Katsaggelos, and Oliver Cossairt, "Computational multifocal microscopy," Biomedical Optics Express 9, 6477-6496 (2018)
